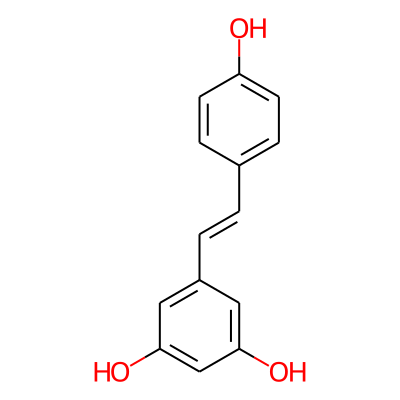
Summary
SMILES: Oc1ccc(cc1)/C=C/c1cc(O)cc(c1)OInChI: InChI=1S/C14H12O3/c15-12-5-3-10(4-6-12)1-2-11-7-13(16)9-14(17)8-11/h1-9,15-17H/b2-1+InChIKey: LUKBXSAWLPMMSZ-OWOJBTEDSA-N
DeepSMILES: Occcccc6))/C=C/cccO)ccc6)O
Scaffold Graph/Node/Bond level: C(=Cc1ccccc1)c1ccccc1
Scaffold Graph/Node level: C1CCC(CCC2CCCCC2)CC1
Scaffold Graph level: C1CCC(CCC2CCCCC2)CC1
Functional groups: c/C=C/c; cO
Chemical classification
ClassyFire Kingdom: Organic compounds
ClassyFire Superclass: Phenylpropanoids and polyketidesClassyFire Class: Stilbenes
NP Classifier Biosynthetic pathway: Shikimates and Phenylpropanoids
NP Classifier Superclass: Stilbenoids
NP Classifier Class: Monomeric stilbenes
Synonymous chemical names:(e)-resveratrol, (z)-1-(3,4-dihydroxyphenyl)-2-(4-hydroxyphenyl)ethylene, 3,5,4'-trihydroxystilbene, 3,5,4-trihydroxystilbene, e-resveratrol, resveratrol, resveratrol ( 3,4',5-trihydroxystilbene), trans-resveratrol
External chemical identifiers:CID:445154; ChEMBL:CHEMBL165; ChEBI:45713; ZINC:ZINC000000006787; FDASRS:Q369O8926L; SureChEMBL:SCHEMBL19425; MolPort-002-499-801
Chemical structure download