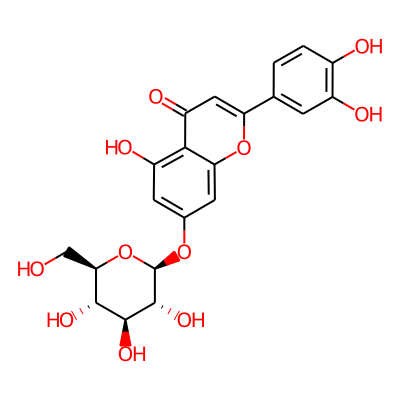
Summary
SMILES: OC[C@H]1O[C@@H](Oc2cc(O)c3c(c2)oc(cc3=O)c2ccc(c(c2)O)O)[C@@H]([C@H]([C@@H]1O)O)OInChI: InChI=1S/C21H20O11/c22-7-16-18(27)19(28)20(29)21(32-16)30-9-4-12(25)17-13(26)6-14(31-15(17)5-9)8-1-2-10(23)11(24)3-8/h1-6,16,18-25,27-29H,7H2/t16-,18-,19+,20-,21-/m1/s1InChIKey: PEFNSGRTCBGNAN-QNDFHXLGSA-N
DeepSMILES: OC[C@H]O[C@@H]OcccO)ccc6)occc6=O)))cccccc6)O))O)))))))))))))[C@@H][C@H][C@@H]6O))O))O
Scaffold Graph/Node/Bond level: O=c1cc(-c2ccccc2)oc2cc(OC3CCCCO3)ccc12
Scaffold Graph/Node level: OC1CC(C2CCCCC2)OC2CC(OC3CCCCO3)CCC12
Scaffold Graph level: CC1CC(C2CCCCC2)CC2CC(CC3CCCCC3)CCC12
Functional groups: CO; c=O; cO; cO[C@@H](C)OC; coc
Chemical classification
ClassyFire Kingdom: Organic compounds
ClassyFire Superclass: Phenylpropanoids and polyketidesClassyFire Class: Flavonoids
ClassyFire Subclass: Flavonoid glycosides
NP Classifier Biosynthetic pathway: Shikimates and Phenylpropanoids
NP Classifier Superclass: Flavonoids
NP Classifier Class: Flavones
Synonymous chemical names:7-glucoside of luteolin, cinaroside, cynaroside, galuteolin, gluco luteolin, glucoluteolin, luteolin -7- glucoside, luteolin 7-glucoside, luteolin 7-o-glucoside, luteolin and its 7 glucoside, luteolin-7-glucoside, luteolin-7-glucosides, luteolin-7-glucosylglucuronide, luteolin-7-mono-beta-d-glucopyranoside, luteolin-7-o-beta-glucoside, luteolin-7-o-glucopyranoside, luteolin-7-o-glucoside, luteolin-7-β-d-glucoside, luteolin-7-β-glucoside, luteoloside
External chemical identifiers:CID:5280637; ChEMBL:CHEMBL233929; ChEBI:27994; ZINC:ZINC000004096258; FDASRS:98J6XDS46I; SureChEMBL:SCHEMBL149118; MolPort-001-740-780
Chemical structure download