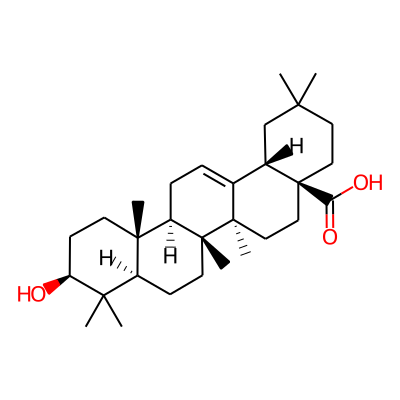
Summary
SMILES: O[C@H]1CC[C@]2([C@H](C1(C)C)CC[C@@]1([C@@H]2CC=C2[C@@]1(C)CC[C@@]1([C@H]2CC(C)(C)CC1)C(=O)O)C)CInChI: InChI=1S/C30H48O3/c1-25(2)14-16-30(24(32)33)17-15-28(6)19(20(30)18-25)8-9-22-27(5)12-11-23(31)26(3,4)21(27)10-13-29(22,28)7/h8,20-23,31H,9-18H2,1-7H3,(H,32,33)/t20-,21-,22+,23-,27-,28+,29+,30-/m0/s1InChIKey: MIJYXULNPSFWEK-GTOFXWBISA-N
DeepSMILES: O[C@H]CC[C@][C@H]C6C)C))CC[C@@][C@@H]6CC=C[C@@]6C)CC[C@@][C@H]6CCC)C)CC6)))))C=O)O))))))))))C)))))C
Scaffold Graph/Node/Bond level: C1=C2C3CCCCC3CCC2C2CCC3CCCCC3C2C1
Scaffold Graph/Node level: C1CCC2C(C1)CCC1C2CCC2C3CCCCC3CCC21
Scaffold Graph level: C1CCC2C(C1)CCC1C2CCC2C3CCCCC3CCC21
Functional groups: CC(=O)O; CC=C(C)C; CO
Chemical classification
ClassyFire Kingdom: Organic compounds
ClassyFire Superclass: Lipids and lipid-like moleculesClassyFire Class: Prenol lipids
ClassyFire Subclass: Triterpenoids
NP Classifier Biosynthetic pathway: Terpenoids
NP Classifier Superclass: Triterpenoids
NP Classifier Class: Oleanane triterpenoids
Synonymous chemical names:3beta-hydroxyolean-12-en-28-oic acid, oleanic acid, oleanolic, oleanolic acid, oleanolic acids, oleanolic-acid, virgaureagenin b
External chemical identifiers:CID:10494; ChEMBL:CHEMBL168; ChEBI:37659; ZINC:ZINC000003785416; FDASRS:6SMK8R7TGJ; SureChEMBL:SCHEMBL71070; MolPort-004-955-261
Chemical structure download