Summary
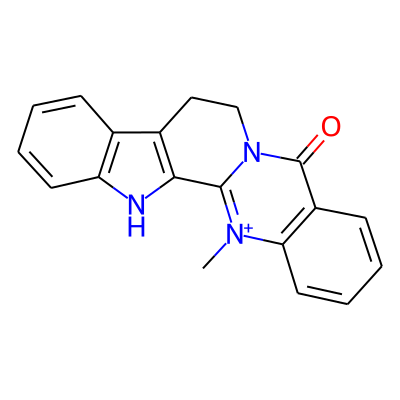
SMILES: C[n+]1c2ccccc2c(=O)n2c1-c1[nH]c3c(c1CC2)cccc3InChI: InChI=1S/C19H15N3O/c1-21-16-9-5-3-7-14(16)19(23)22-11-10-13-12-6-2-4-8-15(12)20-17(13)18(21)22/h2-9H,10-11H2,1H3/p+1InChIKey: VXHNSVKJHXSKKM-UHFFFAOYSA-O
DeepSMILES: C[n+]cccccc6c=O)nc%10-c[nH]ccc5CC9)))cccc6
Scaffold Graph/Node/Bond level: O=c1c2ccccc2[nH+]c2n1CCc1c-2[nH]c2ccccc12
Scaffold Graph/Node level: OC1C2CCCCC2NC2C3NC4CCCCC4C3CCN12
Scaffold Graph level: CC1C2CCCCC2CC2C1CCC1C3CCCCC3CC12
Functional groups: c=O; c[n+](c)C; c[nH]c; cn(c)C
Chemical classification
ClassyFire Kingdom: Organic compounds
ClassyFire Superclass: Organoheterocyclic compoundsClassyFire Class: Indoles and derivatives
ClassyFire Subclass: Pyridoindoles
NP Classifier Biosynthetic pathway: Alkaloids
NP Classifier Superclass: Tryptophan alkaloids
NP Classifier Class: Corynanthe type|Carboline alkaloids
Synonymous chemical names:dehydroevodiamine hydrochloride
External chemical identifiers:CID:156372; ChEMBL:CHEMBL81923; ZINC:ZINC000013434330; MolPort-005-981-197
Chemical structure download