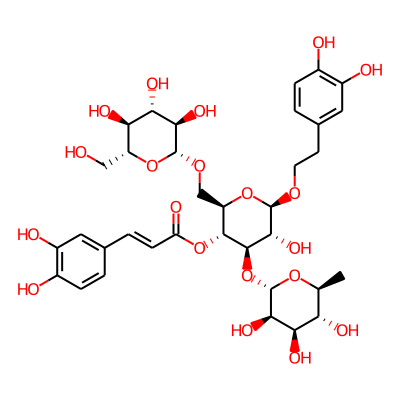
Summary
SMILES: OC[C@H]1O[C@@H](OC[C@H]2O[C@@H](OCCc3ccc(c(c3)O)O)[C@@H]([C@H]([C@@H]2OC(=O)/C=C/c2ccc(c(c2)O)O)O[C@@H]2O[C@@H](C)[C@@H]([C@H]([C@H]2O)O)O)O)[C@@H]([C@H]([C@@H]1O)O)OInChI: InChI=1S/C35H46O20/c1-14-24(42)26(44)29(47)35(51-14)55-32-30(48)34(49-9-8-16-3-6-18(38)20(40)11-16)53-22(13-50-33-28(46)27(45)25(43)21(12-36)52-33)31(32)54-23(41)7-4-15-2-5-17(37)19(39)10-15/h2-7,10-11,14,21-22,24-40,42-48H,8-9,12-13H2,1H3/b7-4+/t14-,21+,22+,24-,25+,26+,27-,28+,29+,30+,31+,32+,33+,34+,35-/m0/s1InChIKey: FSBUXLDOLNLABB-ISAKITKMSA-N
DeepSMILES: OC[C@H]O[C@@H]OC[C@H]O[C@@H]OCCcccccc6)O))O))))))))[C@@H][C@H][C@@H]6OC=O)/C=C/cccccc6)O))O))))))))))O[C@@H]O[C@@H]C)[C@@H][C@H][C@H]6O))O))O)))))))O)))))))[C@@H][C@H][C@@H]6O))O))O
Scaffold Graph/Node/Bond level: O=C(C=Cc1ccccc1)OC1C(COC2CCCCO2)OC(OCCc2ccccc2)CC1OC1CCCCO1
Scaffold Graph/Node level: OC(CCC1CCCCC1)OC1C(COC2CCCCO2)OC(OCCC2CCCCC2)CC1OC1CCCCO1
Scaffold Graph level: CC(CCC1CCCCC1)CC1C(CCC2CCCCC2)CC(CCCC2CCCCC2)CC1CC1CCCCC1
Functional groups: CO; CO[C@@H](C)OC; CO[C@H](C)OC; c/C=C/C(=O)OC; cO
Chemical classification
ClassyFire Kingdom: Organic compounds
ClassyFire Superclass: Organic oxygen compoundsClassyFire Class: Organooxygen compounds
ClassyFire Subclass: Carbohydrates and carbohydrate conjugates
NP Classifier Biosynthetic pathway: Shikimates and Phenylpropanoids
NP Classifier Superclass: Phenylethanoids (C6-C2)|Phenylpropanoids (C6-C3)
NP Classifier Class: Cinnamic acids and derivatives|Phenylethanoids
Synonymous chemical names:echinacoside
External chemical identifiers:CID:5281771; ChEMBL:CHEMBL510539; ChEBI:4745; ZINC:ZINC000085504689; FDASRS:I04O1DT48T; SureChEMBL:SCHEMBL525502; MolPort-001-742-524
Chemical structure download