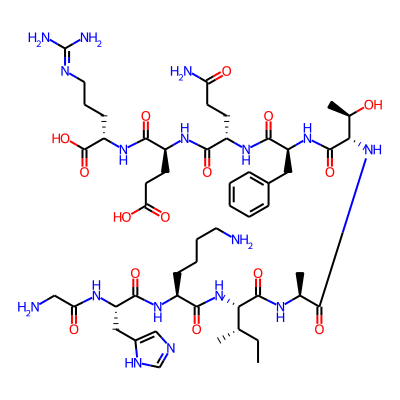
Summary
SMILES: NCCCC[C@@H](C(=O)N[C@H](C(=O)N[C@H](C(=O)N[C@H](C(=O)N[C@H](C(=O)N[C@H](C(=O)N[C@H](C(=O)N[C@H](C(=O)O)CCCN=C(N)N)CCC(=O)O)CCC(=O)N)Cc1ccccc1)[C@H](O)C)C)[C@H](CC)C)NC(=O)[C@H](Cc1[nH]cnc1)NC(=O)CNInChI: InChI=1S/C52H83N17O15/c1-5-27(2)41(68-46(78)32(14-9-10-20-53)63-48(80)37(62-39(72)24-54)23-31-25-58-26-60-31)49(81)61-28(3)43(75)69-42(29(4)70)50(82)67-36(22-30-12-7-6-8-13-30)47(79)65-33(16-18-38(55)71)44(76)64-34(17-19-40(73)74)45(77)66-35(51(83)84)15-11-21-59-52(56)57/h6-8,12-13,25-29,32-37,41-42,70H,5,9-11,14-24,53-54H2,1-4H3,(H2,55,71)(H,58,60)(H,61,81)(H,62,72)(H,63,80)(H,64,76)(H,65,79)(H,66,77)(H,67,82)(H,68,78)(H,69,75)(H,73,74)(H,83,84)(H4,56,57,59)/t27-,28-,29+,32-,33-,34-,35-,36-,37-,41-,42-/m0/s1InChIKey: YWIIIROWEXGMAF-PBEFRTBXSA-N
DeepSMILES: NCCCC[C@@H]C=O)N[C@H]C=O)N[C@H]C=O)N[C@H]C=O)N[C@H]C=O)N[C@H]C=O)N[C@H]C=O)N[C@H]C=O)O))CCCN=CN)N)))))))))CCC=O)O)))))))CCC=O)N)))))))Ccccccc6))))))))))[C@H]O)C)))))C))))[C@H]CC))C)))))NC=O)[C@H]Cc[nH]cnc5))))))NC=O)CN
Scaffold Graph/Node/Bond level: O=C(CCc1cnc[nH]1)NCC(=O)NCC(=O)NCC(=O)NCC(=O)NCCc1ccccc1
Scaffold Graph/Node level: OC(CCC1CNCN1)NCC(O)NCC(O)NCC(O)NCC(O)NCCC1CCCCC1
Scaffold Graph level: CC(CCCC1CCCCC1)CCC(C)CCC(C)CCC(C)CCC(C)CCC1CCCC1
Functional groups: CC(=O)NC; CC(=O)O; CC(N)=O; CN; CN=C(N)N; CNC(C)=O; CO; c[nH]c; cnc
Chemical classification
ClassyFire Kingdom: Organic compounds
ClassyFire Superclass: Organic PolymersClassyFire Class: Polypeptides
NP Classifier Biosynthetic pathway: Amino acids and Peptides
NP Classifier Superclass: Oligopeptides
Synonymous chemical names:yg-1
External chemical identifiers:CID:101594432
Chemical structure download