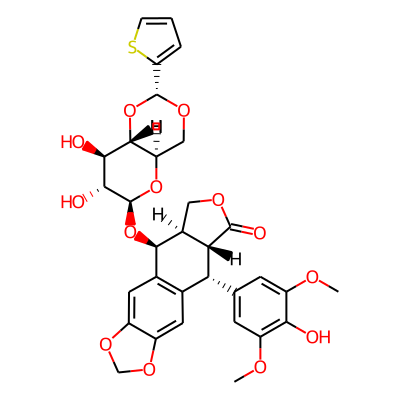
Summary
SMILES: COc1cc(cc(c1O)OC)[C@H]1[C@H]2C(=O)OC[C@@H]2[C@@H](c2c1cc1OCOc1c2)O[C@@H]1O[C@@H]2CO[C@H](O[C@H]2[C@@H]([C@H]1O)O)c1cccs1InChI: InChI=1S/C32H32O13S/c1-37-19-6-13(7-20(38-2)25(19)33)23-14-8-17-18(42-12-41-17)9-15(14)28(16-10-39-30(36)24(16)23)44-32-27(35)26(34)29-21(43-32)11-40-31(45-29)22-4-3-5-46-22/h3-9,16,21,23-24,26-29,31-35H,10-12H2,1-2H3/t16-,21+,23+,24-,26+,27+,28+,29+,31+,32-/m0/s1InChIKey: NRUKOCRGYNPUPR-QBPJDGROSA-N
DeepSMILES: COcccccc6O))OC))))[C@H][C@H]C=O)OC[C@@H]5[C@@H]cc9ccOCOc5c9)))))))))O[C@@H]O[C@@H]CO[C@H]O[C@H]6[C@@H][C@H]%10O))O))))ccccs5
Scaffold Graph/Node/Bond level: O=C1OCC2C(OC3CCC4OC(c5cccs5)OCC4O3)c3cc4c(cc3C(c3ccccc3)C12)OCO4
Scaffold Graph/Node level: OC1OCC2C(OC3CCC4OC(C5CCCS5)OCC4O3)C3CC4OCOC4CC3C(C3CCCCC3)C12
Scaffold Graph level: CC1CCC2C(CC3CCC4CC(C5CCCC5)CCC4C3)C3CC4CCCC4CC3C(C3CCCCC3)C12
Functional groups: CO; COC(C)=O; CO[C@@H](C)OC; c1cOCO1; cO; cOC; c[C@H](OC)OC; csc
Chemical classification
ClassyFire Kingdom: Organic compounds
ClassyFire Superclass: Lignans, neolignans and related compoundsClassyFire Class: Lignan lactones
ClassyFire Subclass: Podophyllotoxins
NP Classifier Biosynthetic pathway: Shikimates and Phenylpropanoids
NP Classifier Superclass: Lignans
NP Classifier Class: Arylnaphthalene and aryltetralin lignans
Synonymous chemical names:vm-26
External chemical identifiers:CID:452548; ChEMBL:CHEMBL452231; ChEBI:75988; ZINC:ZINC000004099009; FDASRS:957E6438QA; SureChEMBL:SCHEMBL3908; MolPort-006-822-656
Chemical structure download