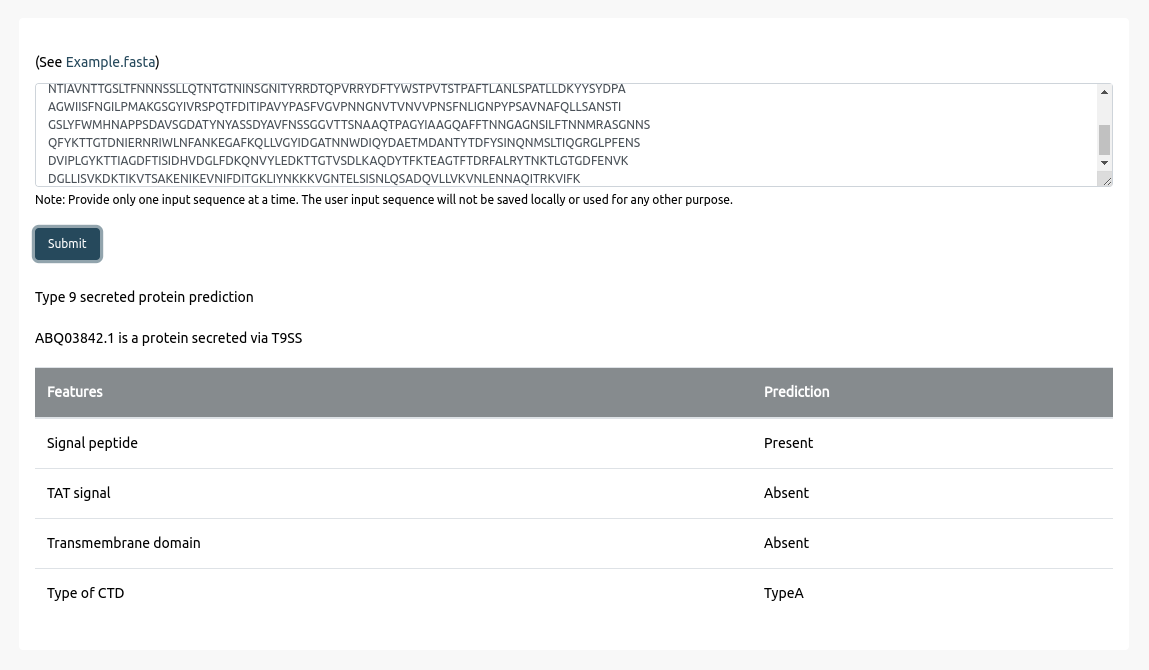
Browse
Browse is an interactive navigation tool providing access to data stored in T9GPred. It tabulates the 693 Bacteroidetes and shows the presence of the 28 protein components in their respective genome.
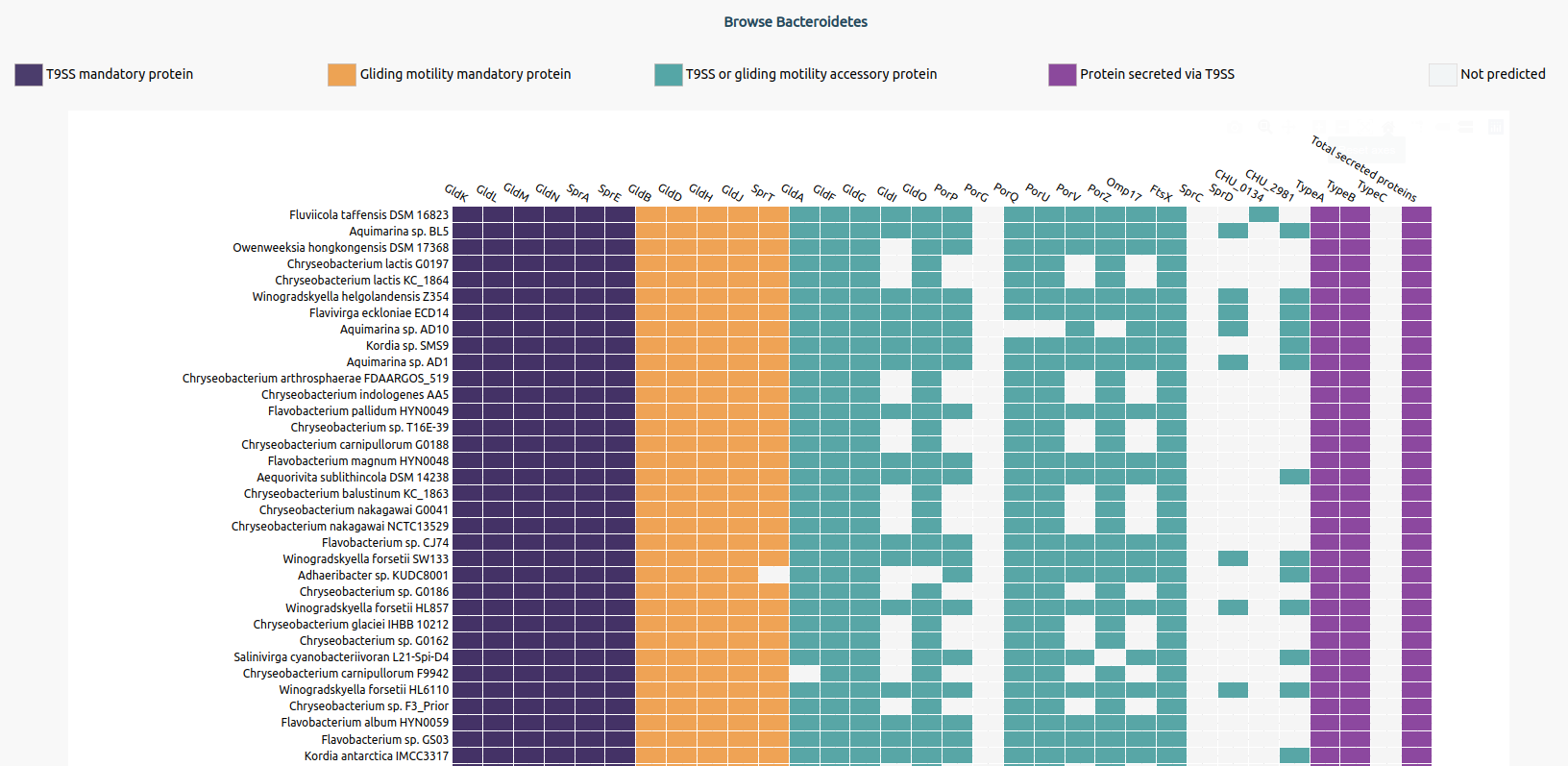
Users can hover over the table to check for Bacteroidetes name, protein component name, and the number of protein sequences predicted for that bacterium.
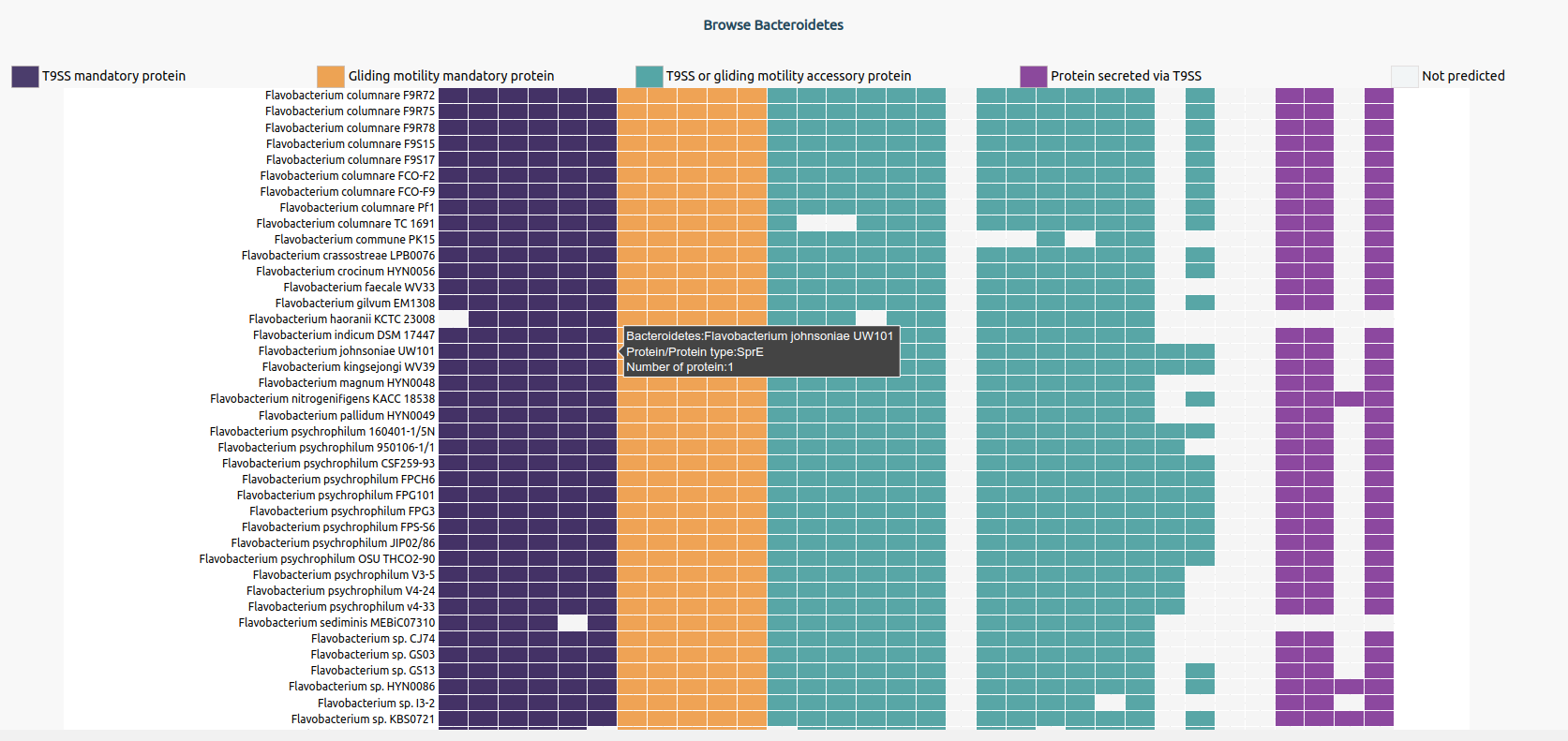
The information about a bacterium can be accessed by selecting the name from the heatmap. Upon selection, the Bacteroidetes information page opens up. It provides information about name, genome identifier, sequencing information, isolation source, predictions, and taxonomy. The proteome of the bacterium can be downloaded by selecting the ‘fasta’ button.
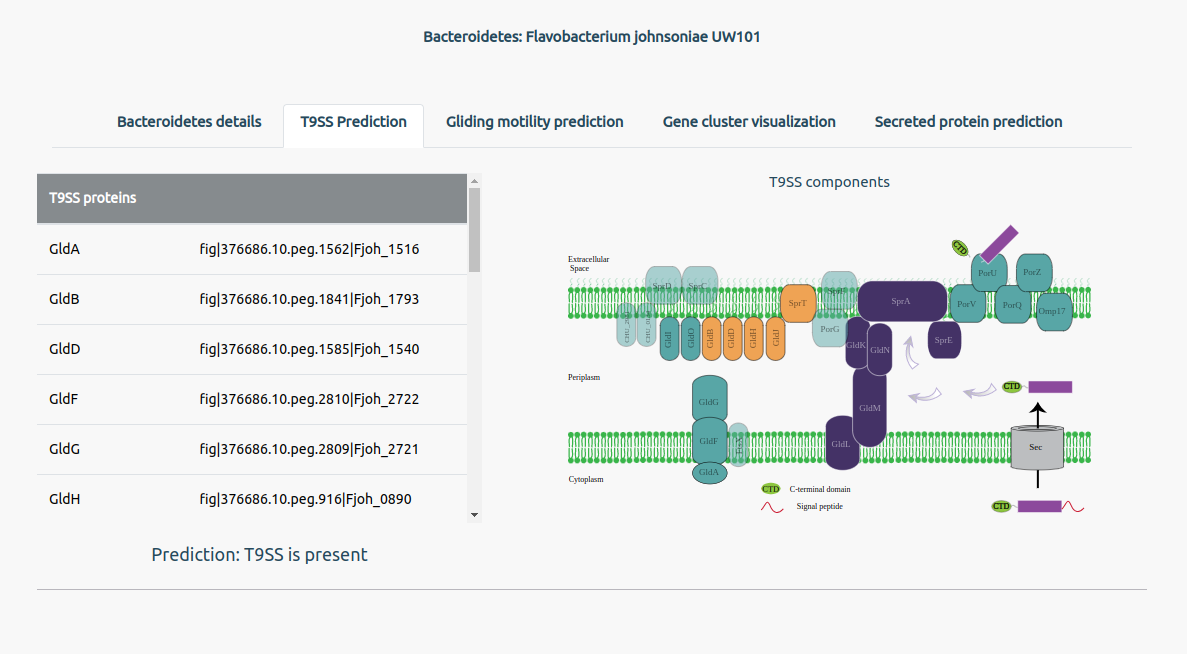
The T9SS prediction tab will give information on the presence of T9SS along with its predicted protein components in the bacterium. The users can also see a visual on the right of the page, highlighting the protein components predicted to be present in the bacterium.
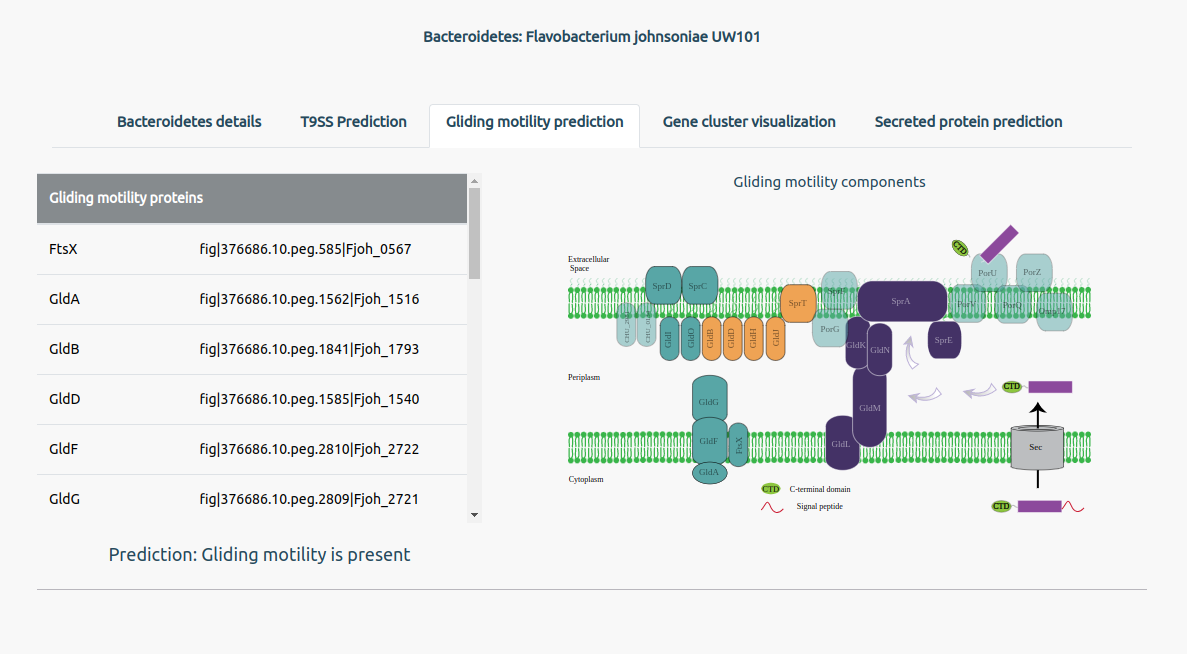
The Gliding motility prediction tab will give information on the presence of gliding motility along with its predicted protein components in the bacterium. The users can also see a visual on the right of the page, highlighting the protein components predicted to be present in the bacterium.
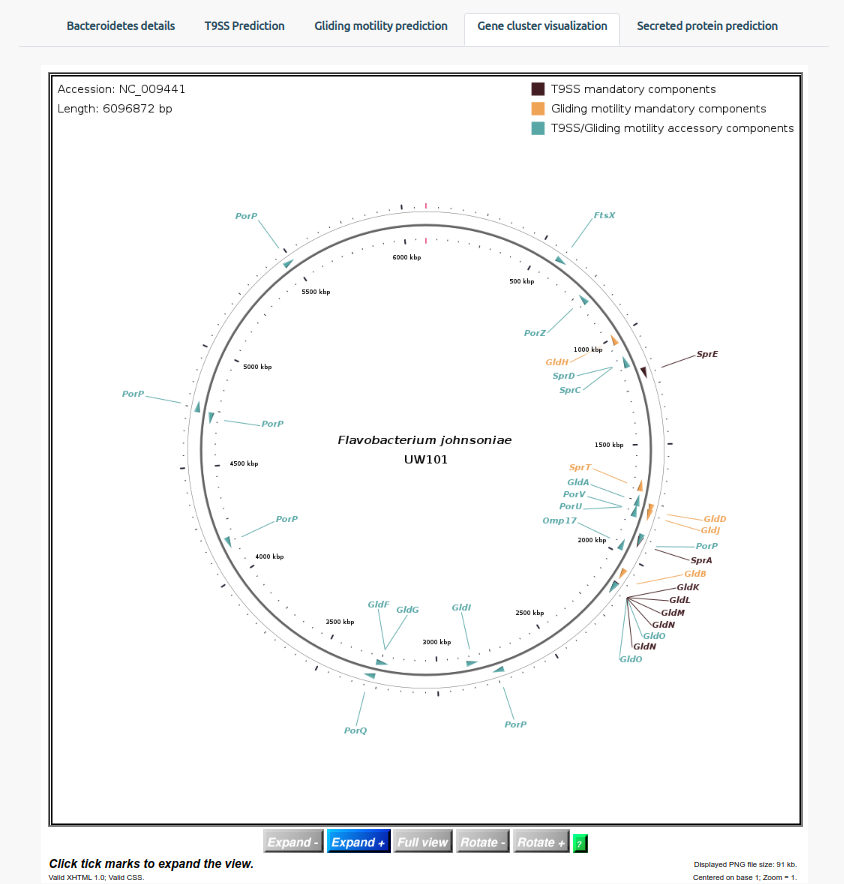
The Gene cluster visualization allows users to visualize the location of the protein components on the genome of the bacterium. Users can zoom in using the ‘Expand+’ option and press ‘Rotate+’ to go through the entire genome.
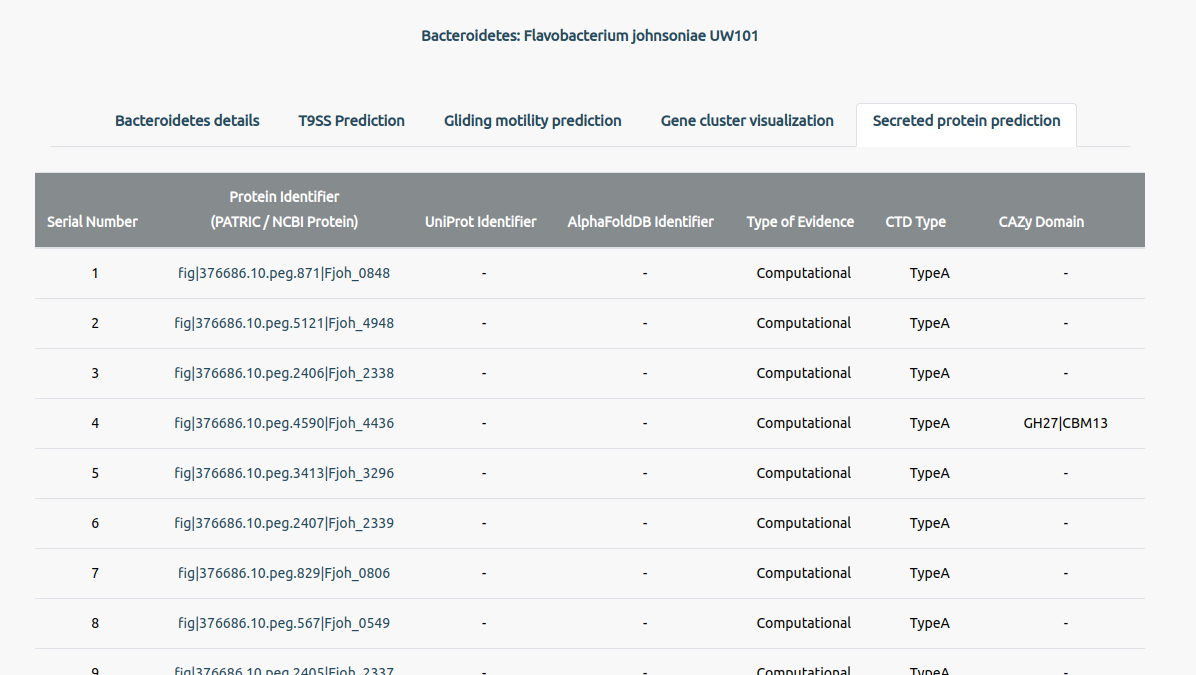
The secreted proteins tab will give information on the predicted secreted proteins in the bacterium (if any). It lists the proteins predicted to be secreted via T9SS along with other information in a tabular format. Users can click on the identifiers to go to their respective databases.
Prediction
Prediction allows users to predict the presence of T9SS or gliding motility in the input proteome and to predict whether the protein can be secreted via T9SS.
- T9SS and Gliding motility prediction
- Protein secreted via T9SS
This tool allows users to predict whether the input proteome contains components associated with T9SS or gliding motility. Users can upload the input proteome in fasta format or copy the proteome onto the text box by choosing the appropriate option.
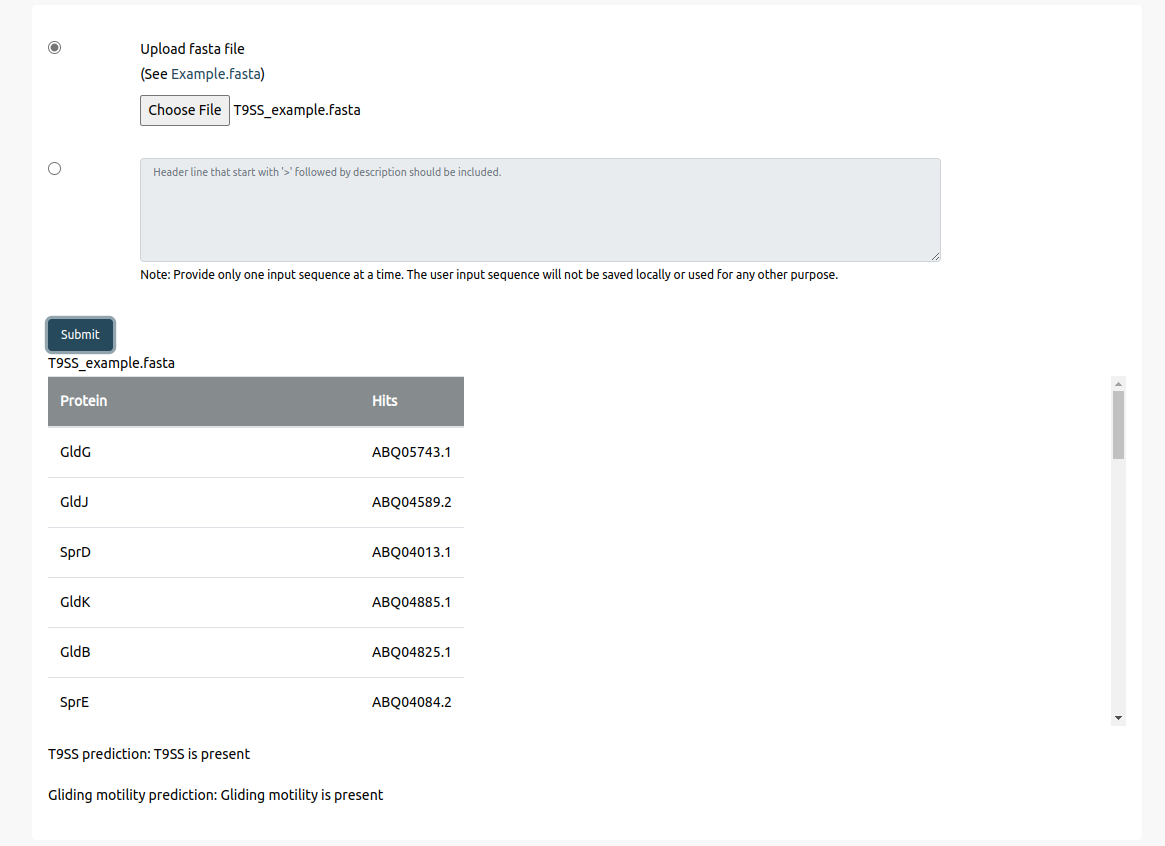
There is an T9SS_Example.fasta file that can be downloaded by selecting 'Example.fasta'. Uploading this fasta file and pressing the ‘Submit’ button will give an output containing the protein components, corresponing hits from the input proteome, and mentions whether T9SS and Gliding motility are present.
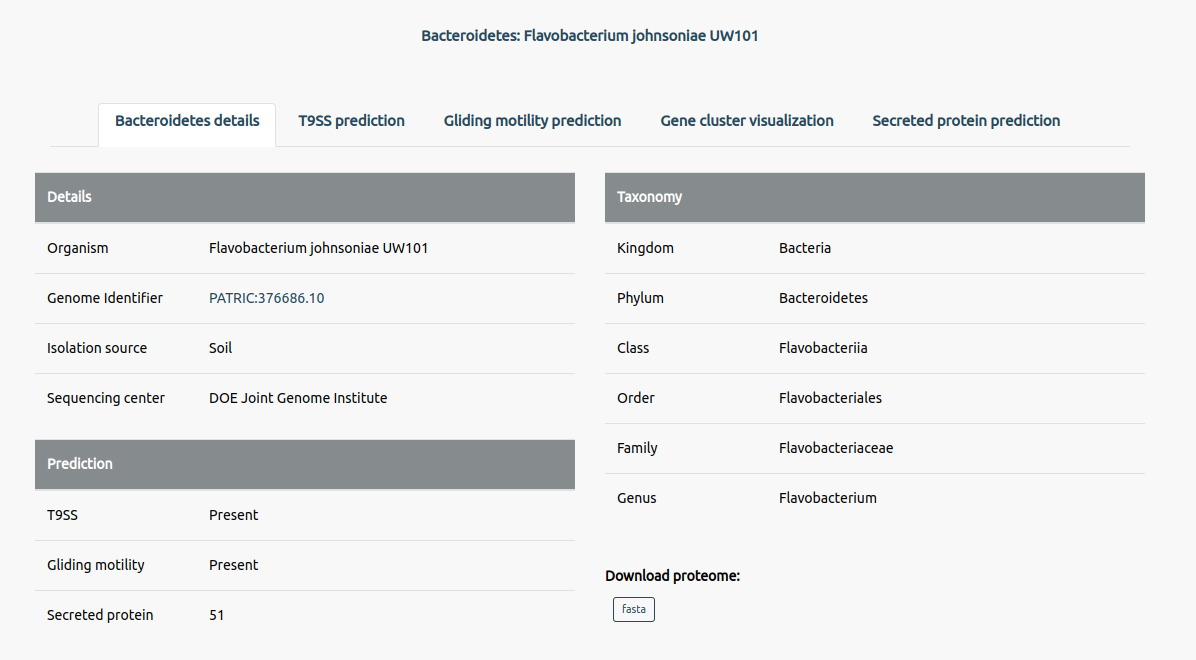
Users can check whether the protein can be secreted via T9SS. Due to computational constraints, users can check for only one protein sequence (maximum of 2048 characters) at a time. To check over multiple sequences, please use the standalone version of the tool available in our GitHub repository.
The contents from the given T9SP_example.fasta file can be used by pressing the 'See Example'. Upon pasting the given sequence in the provided text box and pressing the submit button will yield the output containing the features and their prediction in a tabular format. The tool will mention the possibility of the protein being secreted via T9SS.