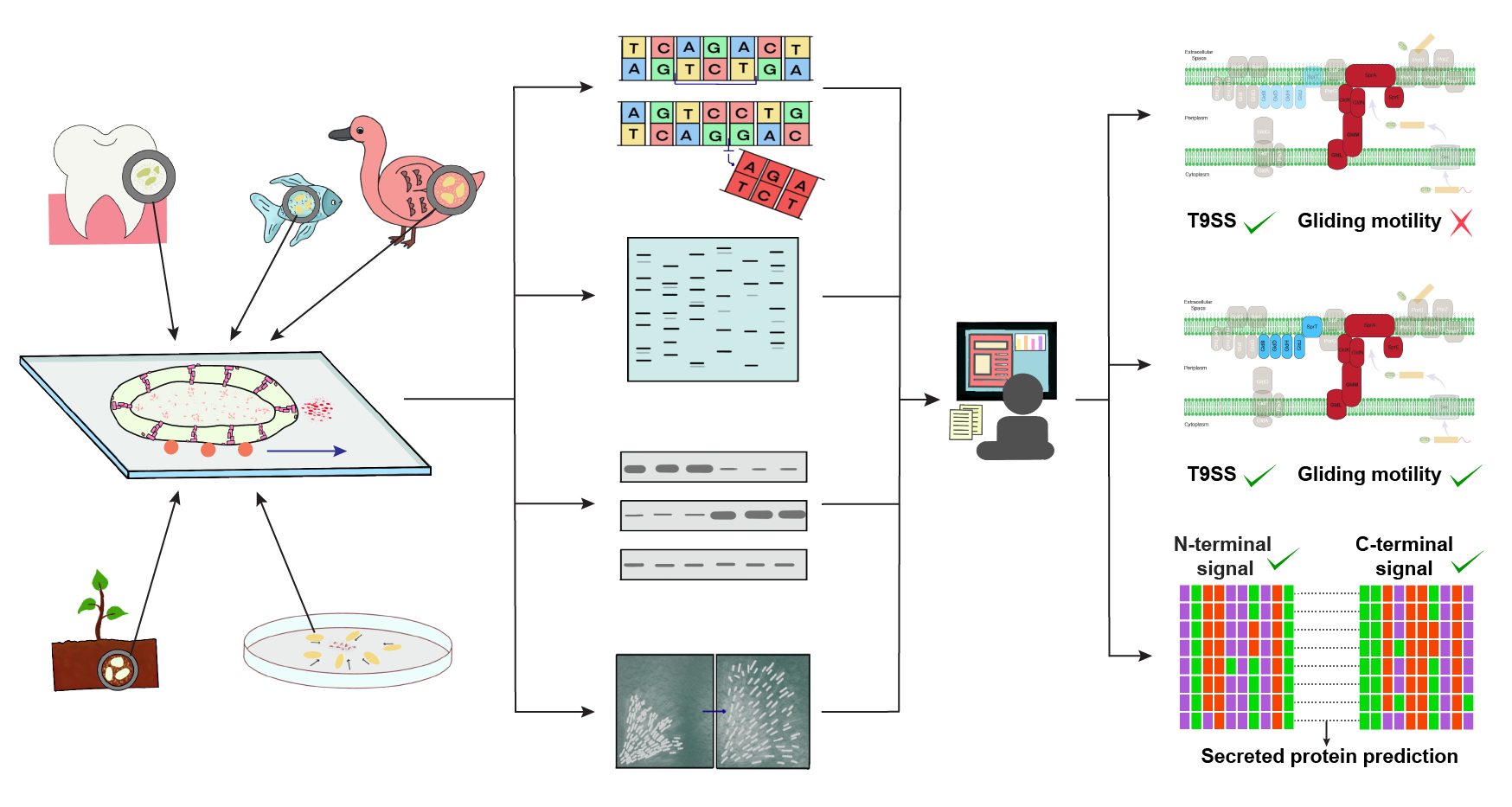
Type 9 secretion system (T9SS) is one of the recently characterized secretion systems found restricted to the Bacteroidetes phylum (environmentally and economically relevant bacterial lineage) [Sato et al (2009), Sato et al (2013), McBride & Zhu (2013)]. T9SS is known to play a central role in the secretion of proteins involved in diverse bacterial functions, including survival, movement and pathogenicity. The bacterial movement associated with T9SS is termed as gliding motility [Sato et al (2009)]. It is interesting to note that not all bacteria having T9SS exhibit gliding motility.
Here, we present a computational tool which predicts the presence of T9SS, gliding motility or proteins secreted via T9SS from proteome of bacteria. We identified 28 proteins associated with T9SS or gliding motility from published literature. From these 28 proteins, we designated 6 proteins (GldK, GldL, GldM, GldN, SprA, SprE) as mandatory components for the presence of T9SS and an additional 5 proteins (GldB, GldD, GldH, GldJ, SprT) as mandatory components for presence of gliding motility. Using this knowledge, we predicted 402 of 693 completely sequenced Bacteroidetes strains to have T9SS. Out of these 402, 327 Bacteroidetes strains are predicted to also have gliding motility. We also visualized the location of the identfied protein components in the genome of respective bacteria. Further, we also predicted the proteins secreted via T9SS across 402 Bacteroidetes strains having T9SS. For the predicted secreted proteins, we provide the protein identifier from PATRIC or NCBI Protein, UniProt identifier, AlphaFold DB identifier, type of evidence, type of C-terminal domain (CTD) and the presence of Carbohydrate-Active enZYmes (CAZy) domain.
The predicted results can be viewed in the Browse section. The computational tool developed here can be accessed in the Prediction section. A standalone version of the tool is available on our GitHub page.
Citation
If you use our resource, please cite the following research article:Ajaya Kumar Sahoo, R. P. Vivek-Ananth, Nikhil Chivukula, Shri Vishalini Rajaram, Karthikeyan Mohanraj, Devanshi Khare, Celin Acharya and Areejit Samal*, T9GPred: A Comprehensive Computational Tool for the Prediction of Type 9 Secretion System, Gliding Motility, and the Associated Secreted Proteins, ACS Omega 8:34091–34102 (2023).
*Corresponding author
Contact
In case you face problems, please contact Areejit Samal.
Funding
Research in the group of Areejit Samal at The Institute of Mathematical Sciences (IMSc), Chennai is financially supported by the Department of Atomic Energy (DAE) India, a Ramanujan fellowship (SB/S2/RJN-006/2014) from the Science and Engineering Research Board (SERB), Department of Science and Technology (DST) India, and a Max Planck Partner Group funded by the Max Planck Society Germany. The funders have no role in study design, prediction, analysis or decision to publish this work.
Disclaimer
T9GPred is an online webserver that predicts the presence of T9SS and gliding motility from an input proteome. T9GPred also predicts proteins secreted via T9SS from an input proteome. Additionally, users can get the prediction by submitting the proteome of an organism. The user input protein sequences are not stored locally and not used for any of our purposes. We are not responsible for errors or omission in published literature or protein databases used to compile the information in this resource.